 |
| Home | Research | Publications | Prospective Students | Teaching | Contact | Physics | Resources | Fun |
| Research Home
Current Projects Other Projects |
Insights into DNA Structure and Dynamics Through Coarse-Grained Models
Modeling bio-molecules at a coarse-grained level of description is often either essential to capture the physics of a process that occurs over long time-scales, or useful because the dynamics of many fine-scale degrees of freedom are irrelevant. On the other hand, modeling complex molecules or parts of molecules with points or spheres, a common practice, is often too extreme an approximation. Several interesting issues arise when one uses ellipsoids to coarse-grain molecular constituents such as the bases in DNA. We have recently developed molecular dynamics routines for handling the singularities that arise when one simulates rotational dynamics using traditional Euler angle approaches. A singularity-free description involves dynamics on a 4-dimensional unit sphere, using quaternions. This quaternion based, molecular dynamics can then be applied to study biomolecular processes in DNA and proteins that occur on long time-scales, such as DNA translocation through a nanopore, or force-induced protein unfolding. 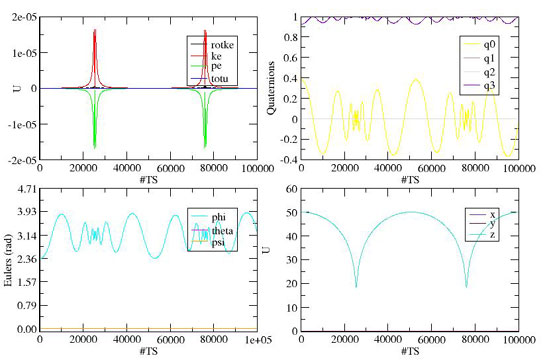
We have introduced a coarse-grained model of DNA with bases modeled as rigid-body ellipsoids to
capture their anisotropic stereochemistry:
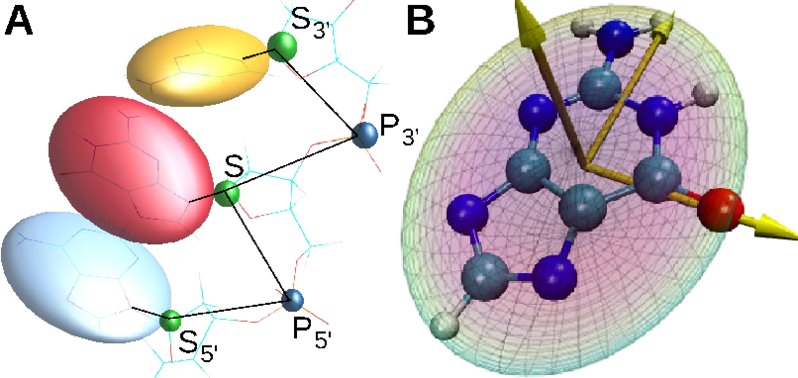
Interaction potentials are all physicochemical and
generated from all-atom simulation/parameterization with minimal phenomenology.
Persistence
length, degree of stacking, and twist are studied by molecular dynamics simulation as functions of
temperature, salt concentration, sequence, interaction potential strength, and local position along the
chain for both single- and double-stranded DNA where appropriate.
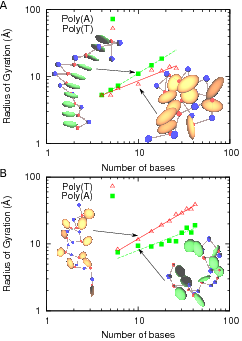
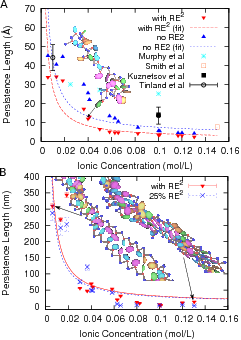
| Radius of gyration of ssDNA for poly(A) and poly(T) as a function of the number of bases, shows different behavior depending on the sequence in two temperature regimes. | Persistence length vs ionic concentration for both single and double stranded DNA. Both experimental and theoretical values are shown for ssDNA. We are able to investigate the role of stacking interactions on the persistence length of DNA by making them stronger or weaker; the results are nontrivial (see our paper below). |
The model of DNA shows several phase transitions and crossover regimes in addition to dehybridization, including unstacking, untwisting, and collapse, which affect mechanical properties such as rigidity and persistence length.
The model also exhibits chirality with a stable right-handed and metastable left-handed helix.
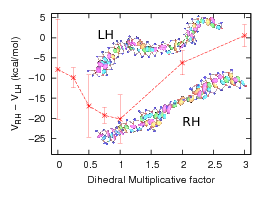
| Difference in the thermal average potential energy between righthanded and left-handed forms of dsDNA. |
References:
[1] Morriss-Andrews, A, Rottler J, Plotkin SS, "A systematically coarse-grained model for DNA, and its predictions for persistence length, stacking, twist, and chirality" J Chem Phys 132, 035105 (2010)
Home | Research | Publications | Prospective Students | Teaching | Contact | Physics | Resources || Fun |