 |
| Home | Research | Publications | Prospective Students | Teaching | Contact | Physics | Resources | Fun |
BioVEC : A program for Biomolecule Visualization with Ellipsoidal Coarse-graining
| BioVEC | Images | Videos | Download | Citation |
BioVEC is a tool for visualizing molecular dynamics simulation data while allowing coarse-grained residues to berendered as ellipsoids. The use of coarse-graining methods in biomolecular simulation is progressively increasing, mainly to circumvent the current time-scale problem that exists for all-atom molecular dynamics simulations, and to allow for the computational modelling of ever larger systems which address biologically relevant questions. However in spite of this progress, to the authors' knowledge no currently available program allows for the straightforward visualization the dynamics of coarse-grained systems with ellipsoidal constituents.
More examples of images created by BioVEC:
 BioVEC running on Windows XP |
 Globule formed by single stranded DNA |
 Thermal anisotropy of GM1-pentasaccharide from a complex with the cholera toxin B-pentamer
Thermal anisotropy of GM1-pentasaccharide from a complex with the cholera toxin B-pentamer |
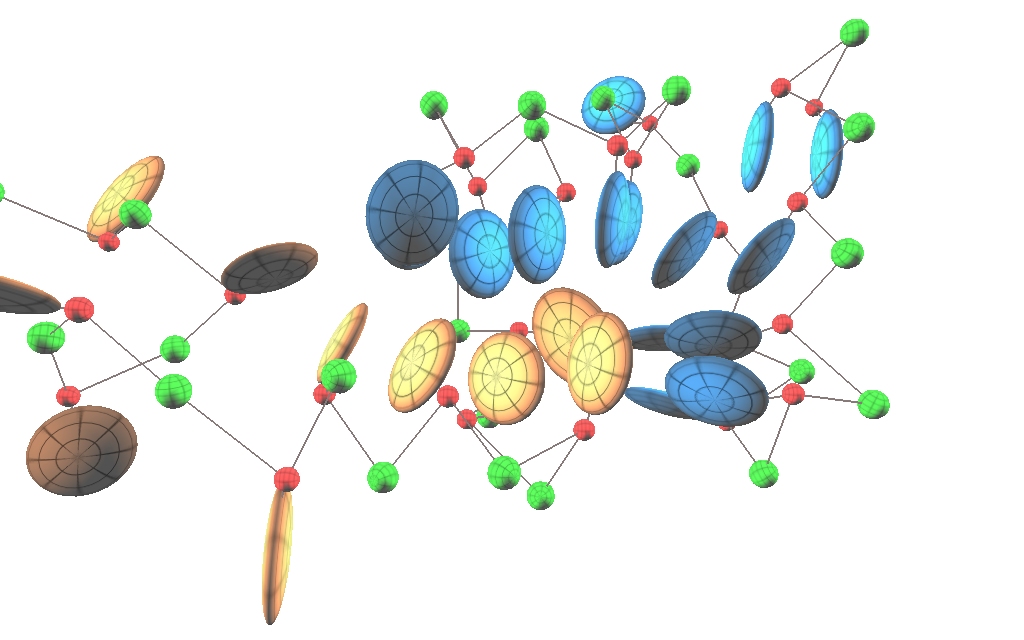
 ANISOU representation of the 1IRO protein
ANISOU representation of the 1IRO protein
|
BioVEC reads in configuration files, which may be output from molecular dynamics simulations that include orientation output in quaternion format, and can render frames of the trajectory in several common image formats for subsequent concatenation into a movie file. Below is shown an example of a single stranded DNA, consisting of two blocks of A and T bases, forming a partial hairpin. The raw simulation data has been averaged over eleven consecutive time steps, to create a smoother animation.
More videos from BioVEC:
 (AVI 14MB) |
 (AVI 14MB) |
Download BioVEC
BioVEC was developed by Erik Abrahamsson while in the Plotkin lab. All source codes and documents are released under the GNU General Public License.- Windows:
- Precompiled Windows executables
- Source code and Dev-C++ Project
- Source code and VC++ 2009 Project
- Windows C++ Runtime Libraries: msvcr80.dll
- Linux:
- BioVEC Source code
- Additional compilation notes (courtesy of Oriol Vilanova, PhD, U. de Barcelona)
- Helper programs:
- Smooth.cpp
- Manual:
- README and documentation
- Example files:
- Input files for BioVEC
- Libraries required to compile BioVEC:
- DevIL
- GNU Scientific Library
- OpenGLUT
- FreeGLUT
- FreeGLUT for VS 2008
- C++ Compilers and IDE for Windows:
- Dev-C++
- Microsoft Visual C++ 2008 Express
BioVEC Citation
Abrahamsson, E. and Plotkin, S. S., ”BioVEC: A program for Biomolecule Visualization with Ellipsoidal Coarse-graining” J. Mol. Graph. Mod. 28, 140-145 (2009) (PDF)
We are pleased to announce that the BioVEC video included in the supplementary materials has won the MGMS Graphics Prize for Best Annual Artwork Submitted to the Journal of Molecular Graphics and Modelling 2010.
Home | Research | Publications | Prospective Students | Teaching | Contact | Physics | Resources || Fun |